Our Development for better Health & Eco Planet

Microbiome and Rapid AMR(Anti microbial Resistance) Detection

Healthcare Services: Monitoring programme

Bulk Chemical Supplies

Next Generation
Advance Battery Research

Healthy Lifestyle Managment Oxidation stress free Food Ingredient
19 Aug 2022
Our Development Summary: ANTIMICROBIAL RESISTANCE AND SURVEILLANCE
The widespread presence of antimicrobial resistance (AMR) is alarming and emerging associated with infection in both hospital and community settings. In regards to antimicrobial resistance, Prof. Dame Sally Davies, the UK Chief Medical Officer (CMO), suggested that AMR should be included on the National Risk Register, due to its potential effect on health. According to challenges emphasized in the 2011 Annual Report of the CMO the necessity to upgrade diagnostic assay for ideal therapeutic interventions through point-of-care (POC) testing. (Davies et al., 2013) In regard to AMR, (Varadi et al., 2017) studies projected that around 10 million of annual global deaths will be the cause of AMR by 2050, if they are not tackled which will contribute more deaths than the death caused by cancer, which is around 8.2 million. This incidence of drug resistance can be reduced by the administration of the right antibacterial agent for the right patient employing pathogen-directed therapy (Davies et al., 2013). Conversely, in developed countries the 90% of human antibiotic use can be adopted only with the prescription in general practise along with the POC test to boost the treatment outcome of patients by minimising the probability of developing resistance in organisms by injudicious use of antibacterials. Prescriptions will be mandatory for the administration of antibiotics and also for the testing technology by 2020, in concern with the UK review on antimicrobial resistance, chaired by Lord O’ Neill (Váradi et al., 2017). In order to prevent the spread and development of drug resistance of bacteria, US National Institutes of Health (NIH), National Institute of Allergy and Infectious Disease (NIAID) funded $20m Antimicrobial Resistance Diagnostic Challenge to discover the quick and novel POC diagnostic assay (Váradi et al., 2017). For an instance,25 000 deaths p.a., has been estimated due to resistant organisms with a productivity loss and healthcare cost of €1.5 billion in Europe whereas 23 000 deaths and 2 million illness were estimated in US associated with the antibacterial resistance, approximately $30 billion has been estimated to spend for both medical and prolonged hospital care. As stated by (Váradi et al., 2017), Enterococcus faecium, staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa and Enterobacter species contributes majority of nosocomial infection and their ability to interrupt and escape from the working of antibacterial agents due to their MDR property are termed as ESKAPE. According to the Centers for Disease Control and Prevention (CDC), 250 000 infections were reported with the clostridium difficle with 14 000 deaths per annum with a medical cost of 1 billion which made them as top level of emerging threat in the U.S.A. carbapenem-resistant Enterobacteriaceae exhibits resistance to almost all antibiotics except aminoglycosides, tigecycline, Fosfomycin and colistin which are the last line antibiotics. The incidence of infection with CRE was reported as 9000 per annum. (Bassetti, Merelli, Temperoni and Astilean, 2013) The identification and characterization of pathogenic bacteria and their susceptibility towards antibiotics have a great impact in antibacterial therapy. To employ detection of pathogenic bacteria in any global settings, an ideal identification method should be simple and easy to handle with no use of sophisticated instruments and uncomplicated result interpretation. It should be rapid by providing results with a time frame, reliable and should be cost effective. Surveillance plays a vital role in the demonstration of accurate and updated information about wide trends with sufficient details regarding new infections and resistance by analysing incidence of cross infection, epidemiological data and government-enabled networks and it becomes more effective when it is comprehensive and communicated. All these data were gathered from the surveillance programme set up by USA (Active Bacterial Core surveillance, ABCs) in association with CDC in 1995 and EU (European Centre for Disease Prevention and Control, ECDC in 1996 (Váradi et al., 2017). Apart from that, in 2014 World Health Organisation (WHO) conducted a study on global antibacterial surveillance with an observation of inconsistency in surveillance across many parts of the world. (Váradi et al., 2017), The rise in the incidence of 3 rd generation cephalosporin and fluoroquinolone resistant E. coli and 3 rd generation cephalosporin resistant K. pneumoniae, and notable rise in the incidence of carbapenem resistant K. pneumoniae was observed all over the world nevertheless of the international effort.
Due to these impacts further surveillance programmes were initiated, current studies show that still some countries lack surveillance programmes with limited standard and international coordination. By gathering data from all over the world The Center for Disease Dynamics Economics and Policy (CDDEP) Resistance Map regarding antimicrobials and resistance in a variety of forms of interrogation. By reviewing their data of CDDEP’s by ‘The State of the World’s Antibiotics 2015’, encouraged countries without antibiotic policy to initiate new national policies through antibiotic stewardship and the conclusions of CDDEP’s Global Antibiotic Resistance Partnership (GARP) were slice up to six essential strategies for national antibiotic policy (The State of the World’s Antibiotics, 2015 – Center for Disease Dynamics, Economics & Policy (CDDEP), 2021). Accurate identification and detection of pathogenic bacteria along with the high sensitivity, specificity, turn-around time, direct detection of bacteria from clinical samples and right drug administration by the evaluation of antibiotic susceptibility testing are the two salient features of surveillance. According to (Franco-Duarte et al., 2019), the genetic features and biochemical abilities are the two factors that determine the pathogenicity of strains and the potential use of microorganisms and suggested that characterization and identification followed by taxonomical classification of biological material will aid to treat infection in near future. Identification and systematics are closely related to each other. Although relationships, possible interactions, diversity of the organism comprises the systematics whereas the classification of organisms in hierarchical structure contributes the taxonomy.
For instance, the mostly used hierarchical classification involves, kingdom, division, class, family, genus, species and followed by identification of strain. Ideal method for the identification of bacteria can be achieved by blending the conventional methods with the molecular techniques. Foundation of research on infection is the isolation of organisms as stated by Robert Koch. Application of Koch’s postulates aids the relationship between bacteria and specific disease. The identification and characterization of bacteria associated with specific diseases paved the way for the advances in pathogenic bacteriology. The clinical samples were inoculated to selective media to obtain pure cultures, the obtained isolates were undergone microscopy by Gram stain and the morphology of the isolated-on agar plates were observed and their characteristic features were noted and followed by catalase and oxidase test obtain their genus identification (e.g., staphylococcus or streptococcus) of the bacteria were carried out. For the species level identification, the isolates underwent specific biochemical or serological tests (latex agglutination test), because the pathogenicity, resistant profile, antibiotic treatment, severity of infection varies between different species within a genus. The incidence of resistance acquired by specific species within a genus exhibits different antibiotic susceptibility profiles. To provide the right drug and proper diagnosis of the treatment plans, the results obtained should be with high level assurance. Moreover, the test methods for the identification of bacteria should be more specific, sensitive, and accurate to detect the antimicrobial susceptibility along with the bacterial genus and species identification (Lagier et al., 2015).
Obtaining genomic data by use of DNA detection is considered as one of the Rapid and highly specific tests for the identification of organisms. The application of DNA Sequencing or PCR (polymerase chain reaction) has transformed gene-based assay, overcome the crisis to identify the infection caused by which bacterial origin and presence of any antimicrobial resistance by the amplification/sequencing Microbiome Genomic material present in the clinical specimen and aids the of rapid testing of samples in a POC healthcare setting . The routine use of genomic testing is expensive, but still, it remains cost effective by ensuring high confidence results. On the other hand, one of the drawbacks of genomic data is its sensitivity. They can identify even a small number of bacteria, below the infection threshold, which leads to initiation of unjudicial antibiotic therapy. The selection of test methods for identification varies with each susceptible pathogen with suitable sensitivity.-
Author- Roshni Philomine (MSc Bio Medical Sciences, Nottingham Trent University), Biomedical Scientist Hasu Diagnostics
19 Dec 2022
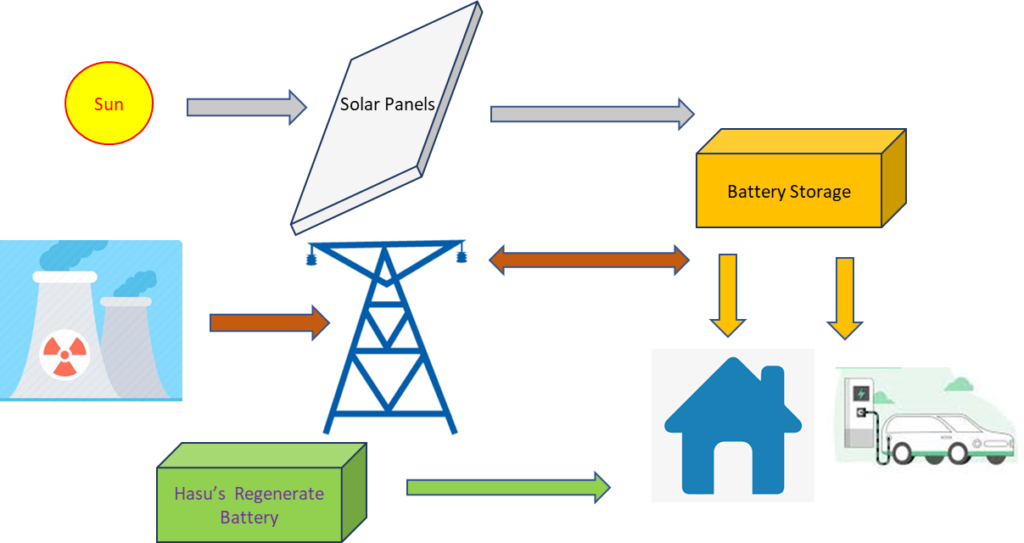
Our Start-up Story: New Generation Energy storage Battery
The Company Founder, Dr. Alpesh Patel, has been passionate about electrochemistry all his life. During his childhood, he was experimenting countless times, with electrochemistry to light the bulbs, or electrochemistry to generate the disinfectant solution through sodium chloride electrolysis and chlorine generation which is absorbed into the caustic soda solution to make the hypo solution.
Even in his study during Years 5 to Year 7, he was triggering a Casio digital watch to be powered with a zinc/copper cell run by a potato. Eventually, one day when he had sourced some very concentrated sulfuric acid to make the voltaic cell at home, he got into an accident and was left with an acid burn on his chest. Thereafter he was banned temporarily by his parents from doing any experiments.
Even though he kept himself busy and focused on his further studies, he acquired some very interesting and amazing experience working with the Indian Petrochemicals R&D centre for synthesising the catalysis for olefin polymerisation. This incredible experience allowed him to work with very sensitive chemicals and metal complexes under an inert atmosphere. He carried on his intensive research and gained a Ph.D. in polyolefin polymers from the MS Univeristy of Baroda .
Whilst studying his main Ph.D. subject, he also worked on other subjects, such as developing various novel chemistry and Innovation e.g inert atmosphere catalyst synthesis for atom transfer polymerisation, and novel amphiphilic cationic surfactant.
Since after PhD and Post doctorate from the Imperial College London he has spent several years of spending his career in molecular diagnostics, he came up again with an idea, to solve the electrochemistry for storage of energy in the form of electric emf. This will potentially solve the greater problem our governments and businesses are facing in developing next-generation, eco-friendly sustainable automobiles, and as a result, revolutionize the energy market.
We, as humans, have unfortunately inflicted enough damage on this beautiful Earth causing a climate crisis and countless species to be extinct. Through our over-ambitious nature, we have pumped out billions of barrels of crude oil and extracted tonnes of coal from the Earth, and converted them into CO2 in the Earth’s atmosphere. This is now making all forms of life difficult to exist and there is increasing and bigger irreversible damage to all our plants, micro-organisms, and all other living organisms.
The battery concept Dr. Alpesh Patel is working on is to innovatively change the highly sensitive electrochemistry, incorporating a novel application of cell regeneration methodology, instead of entirely depending on the grid or solar energy for regeneration or recharging of the battery.
15 Dec 2022